Weber Lab
Genetics, ecology, and evolution of adaptive traits
Strategy
The lab's research explores the process of adaptation in animals.
Specifically, "how" and "why" do animals evolve complex
traits to manage environmental pressures? To answer
these questions, we draw from tools and theory associated with
the fields of:
-
behavioral ecology
-
bioinformatics and genomics
-
biomechanics
-
ecology
-
evolutionary biology
-
immunology
-
molecular and quantitative genetics
All projects employ the following strategy:
1) Identify traits that vary in ecologically meaningful ways
2) Characterize the locus (or loci) of evolution
- Identify the chromosomal regions (and lists of genes/mutations)
- Describe molecular and functional differences between alleles
- Experimentally manipulate interesting candidate genes in the lab
3) Test how alleles affect fitness in the wild
- Experiments that manipulate both genetic background and environment
- Comparative approaches that test for correlations between genetic changes and
environmental variation, using data from many populations and species.
- Long-term field studies that measure allele frequency fluctuations across
many generations.
Host-parasite coevolution
Parasites evolve numerous strategies to manipulate and evade host defenses. And depending on the the cost of infection, hosts can evolve to resist or tolerate parasites. This reciprocal antagonism can drive rapid evolution (hence the Red Queen cartoon from Through the Looking Glass). Although there are numerous population genetic models that aim to explain how this process might proceed, they all rely on assumptions about how host and parasite genetic variation are connected to infection success. At present, there is limited empirical data to test these models, especially with respect to vertebrate hosts. My work aims to fill this gap, and to answer
fundamental questions such as:
What genetic changes underlie host-parasite coevolution?
How often are new genes or strategies deployed, as
opposed to recycling existing variation?
How does gene flow among many populations impact the
rate and trajectory of (co)evolution?
What ecological conditions lead to quantitative variation in
selection, and how does this variation affect the genetic
architecture of adaptations?

Credit: Sir John Tenniel
Sticklebacks and tapeworms:
A powerful model for coevolutionary genetics
1) Plentiful natural diversity
Marine threespine stickleback (Gasterosteus aculeatus) are highly susceptible to Schistocephalus solidus
tapeworms, but freshwater populations across the globe have evolved to resist the parasite. Interestingly, freshwater fish in different areas evolved resistance via different mechanisms, and parasites evolved to counter the resistance of their local hosts (Weber et al 2016).
3) Excellent genomic resources
The threespine stickleback genome is both sequenced and annotated (Ensembl). A draft genome of the parasite is available at ParaSite.WormBase.org.

2) Lab-based infections
We can replicate the parasite's complex life cycle in
the lab (see right figure). This allows us to perform
thousands of controlled exposures between different
host and parasite populations.
We measure numerous
traits in fish and cestodes
after infection.
Breeding cestodes (by crossing or selfing in nylon bags and growth media) produces millions of eggs.
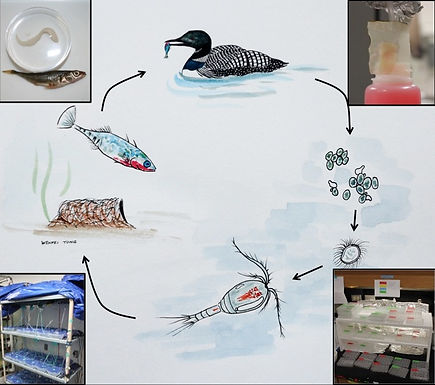
Tapeworm life-cycle:
wild and lab
Eggs hatch in incubated
petri dishes.
Painting credit: Wenfei Tong
Fish are placed in tupperware
and fed infected copepods.
Copepods are reared and exposed to hatched parasites.
4) MANY interesting traits
The pictures below show traits that differ between fish from a High-infection Lake (H) and a No-infection Lake (N), as well as data on their hybrid offspring (Weber et al (2017) PNAS)
Cestode mass: an extended phenotype
of stickleback hosts
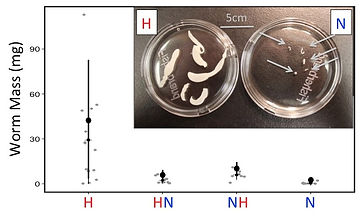
Parasite mass depends on host genetics.
Innate immune responses:
Oxidative burst
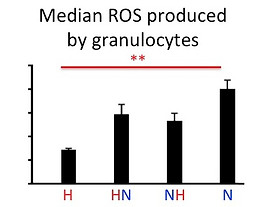
Granulocytes evolved to produce different amounts of Reactive Oxygen Species (ROS).
Ongoing work
Strength of association
with trait variation
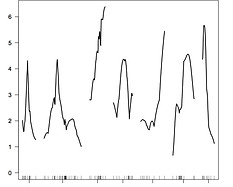
QTLs for
ROS
Stickleback chromosomes
I recently exposed >700 F2-generation hybrid fish (between no-infection and high-infection lakes) to cestodes. Many
genomic regions are strongly associated with infection and/or immune traits, including ROS response (left graph).
We are using multiple approaches (including CRISPR-Cas9 and RNAseq) to test whether particular genes and mutations cause these quantitative trait loci (QTLs).
Adaptations of deer mice (genus Peromyscus)

Peromyscus is a genus of new world mice containing more than 50 species that exhibit astounding levels of diversity. These mice have adapted to most
terrestrial habitats in North America, with some species diversifying into numerous ecotypes. This extreme diversity has long interested biologists, and recent studies have started to dissect the genetic, physiological, and environ-
mental basis of adaptive differences. For example, I collaborated with Drs. Cynthia Steiner and Hopi Hoekstra to identify which genes evolved to create extremely light coat color in beach mice (P. polionotus leucocephalus; image to right), which helps them match the substrates on which they live. But we have only barely started to tap Peromyscus evolutionary genetics gold mine!



My PhD projects focused on the evolution, genetics and ecology of burrow variation in Peromyscus. Initially, I identified some large differences and general trends in how burrowing and nesting vary among species (left figure). I then went on to show that the complex burrows of P. polionotus result from two genetic modules: one set of behaviors control tunnel length, and a separate set influences the construction of escape tunnels (bottom right figure, Weber et al 2013). Through this work I became familiar with the behavior and ecology of numerous
species, and am eager to apply this knowledge in several new lines of research. In particular, I would like to look at the evolution of habitat and diet preferences,
and also connect these to differences in immune traits such as parasite resistance.
Sexual selection manifests itself in many forms, including the extraordinarily large appendages that males use to defend territories and secure access to females. These structures range from the well-known antlers and tusks found in numerous mammals, to the enormous namesakes of stalk-eyed flies. While it is clear that large weapons are likely to both benefits and costs to their owners, there are few case studies that test ecological, genetic, and developmental predictions about what causes weaponary arms-races to escalate or fall apart.

Evolution of Weapons and Combat
Credit: Joi Ito, wikipedia.org
Rhinoceros beetles as a model for sexually selected weapons
As a postdoc in the Emlen Lab, I am examining the evolution of horn length in the Japanese rhinoceros beetle (Trypoxylus dichotomus, photo upper right). The species is distruted throughout China, Taiwan, the Korean peninsula, and Japan. In some populations, males develop large horns that they use to defend sap sites on hardwood trees, where female beetles frequently feed. But in other populations males develop relatively small horns. Females lack horns in all populations. The Emlen Lab and collaborators have already amassed a large knowledge about the developmental processes that give rise to horns. We are now extending this work by sequencing thousands of genetic loci and building a phylogeny from a geographically and morphologically diverse sample of rhino beetles (figure below). We are also examining behavioral variation between the morphs in both field and lab settings. Together, this
will enable us to answer these questions:
How many times has horn length increased or decreased?
What ecological conditions drive
increases or decreases in horn length?
What genes and developmental pathways influence horn evolution?
Are there costs or constraints to horn size?
Do combat-related behaviors evolve in concert with horn evolution?
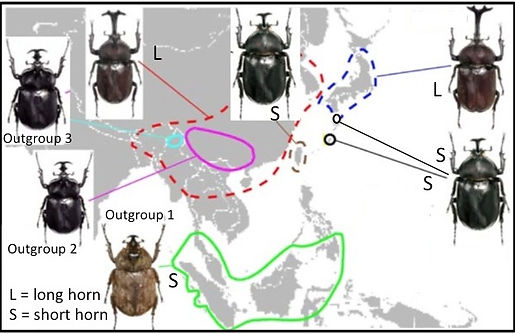
Region = Eastern asia. S = small horn, L = large horn.
Figure credit: Profs. Kunio Araya and Doug Emlen
